SHADE: Modeling Asymmetric Spatial Interactions in Tumor Microenvironments
Summary
Understanding spatial interactions between cell types is essential for interpreting immune function, tumor progression, and tissue organization. While conventional spatial statistics often assume symmetric interactions and analyze images independently, biological systems like the tumor microenvironment frequently exhibit asymmetric, hierarchically structured spatial patterns. We introduce SHADE (Spatial Hierarchical Asymmetry via Directional Estimation), a Bayesian modeling framework that captures direction-specific cell-cell associations across multiple spatial scales using smooth interaction curves and multilevel inference.
Workflow
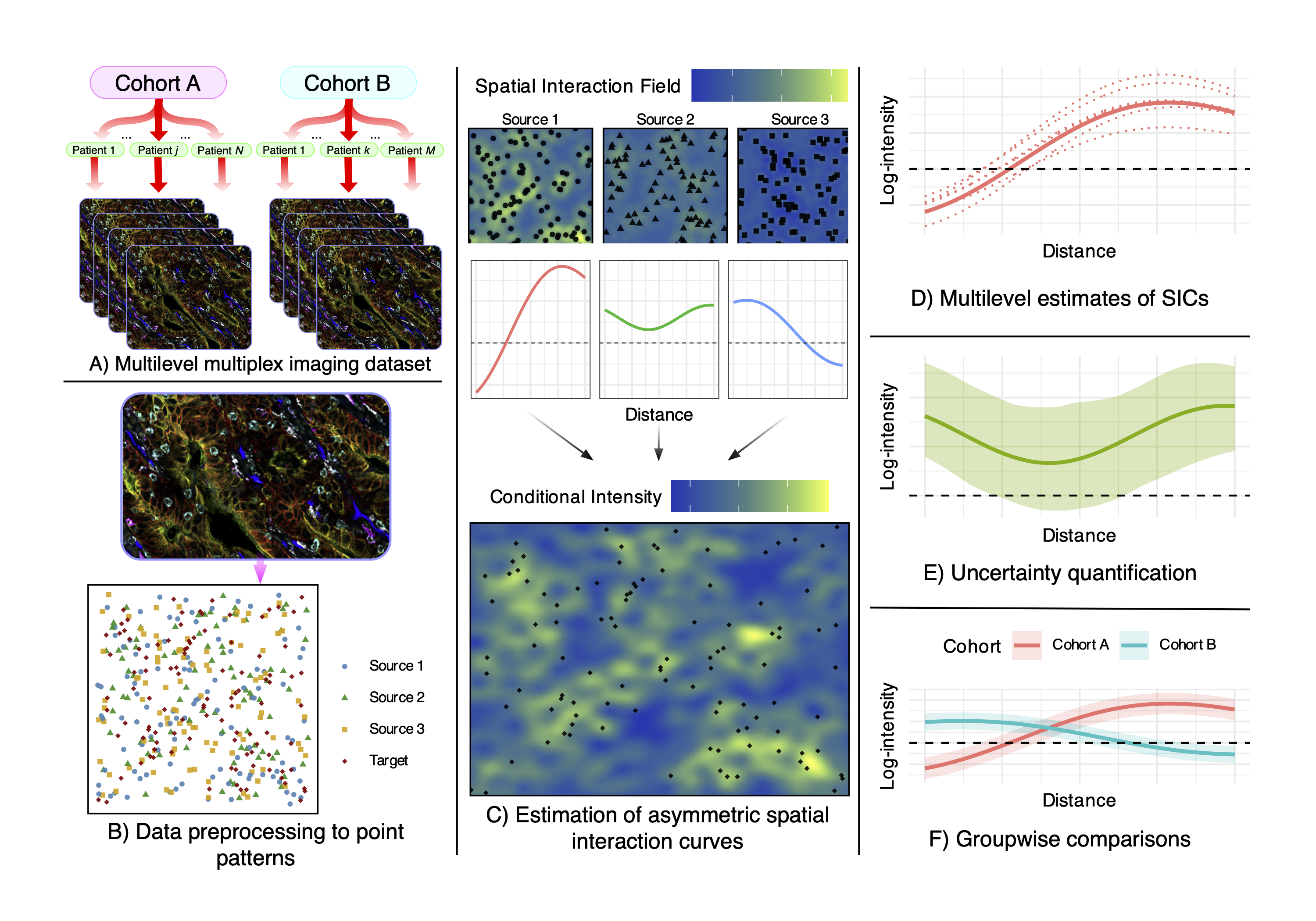
The workflow begins with hierarchically structured image data (cohorts → patients → images), which are processed into annotated spatial point patterns. SHADE then estimates Spatial Interaction Curves (SICs) to capture direction-specific cell-cell associations at multiple biological levels, allowing for analysis of spatial heterogeneity and uncertainty. These estimates can be compared across cohorts to identify differences in spatial organization.